Customize your Processing Pipeline#
This example can be referenced by citing the package.
While NeuroKit is designed to be beginner-friendly, experts who desire to have more control over their own processing pipeline are also offered the possibility to tune functions to their specific usage. This example shows how to use NeuroKit to customize your own processing pipeline for advanced users taking ECG processing as an example.
# Load NeuroKit and other useful packages
import neurokit2 as nk
import numpy as np
import pandas as pd
The Default NeuroKit processing pipeline#
NeuroKit provides a very useful set of functions, *_process() (e.g. ecg_process(), eda_process(), emg_process(), …), which are all-in-one functions that cleans, preprocesses and processes the signals. It includes good and sensible defaults that should be suited for most of users and typical use-cases. That being said, in some cases, you might want to have more control over the processing pipeline.
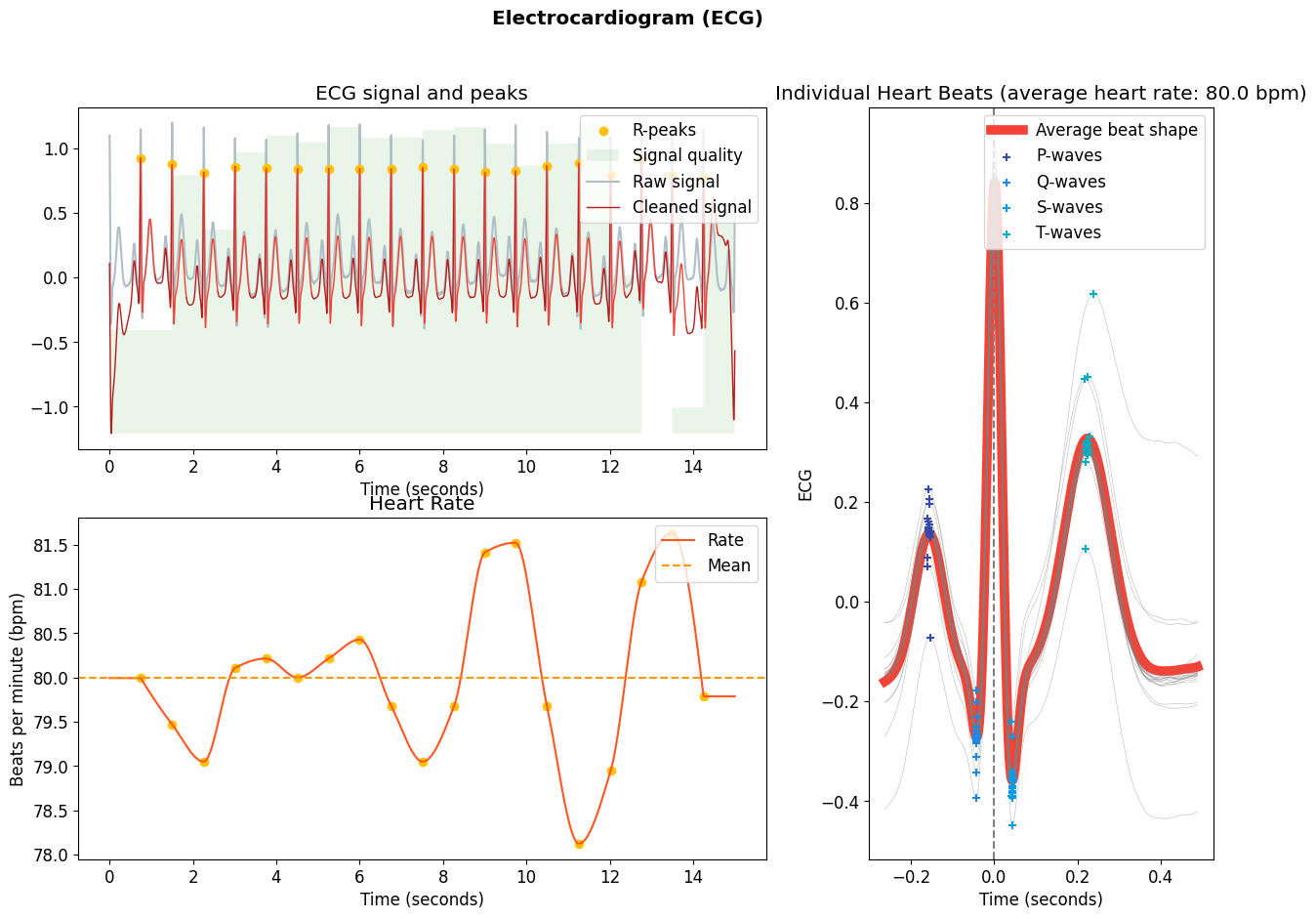
This is how ecg_process() is typically used:
# Simulate ecg signal (you can use your own one)
ecg = nk.ecg_simulate(duration=15, sampling_rate=1000, heart_rate=80)
# Default processing pipeline
signals, info = nk.ecg_process(ecg, sampling_rate=1000)
# Visualize
nk.ecg_plot(signals, info)
Building your own process() function#
Now, if you look at the code of ecg_process() (see here for how to explore the code), you can see that it is in fact very simple.
It uses what can be referred to as “mid-level functions”, such as ecg_clean(), ecg_peaks(), ecg_rate() etc.
This means that you can basically re-create the ecg_process() function very easily by calling these mid-level functions:
# Define a new function
def my_processing(ecg_signal):
# Do processing
ecg_cleaned = nk.ecg_clean(ecg, sampling_rate=1000)
instant_peaks, rpeaks, = nk.ecg_peaks(ecg_cleaned, sampling_rate=1000)
rate = nk.ecg_rate(rpeaks, sampling_rate=1000, desired_length=len(ecg_cleaned))
quality = nk.ecg_quality(ecg_cleaned, sampling_rate=1000)
# Prepare output
signals = pd.DataFrame({"ECG_Raw": ecg_signal,
"ECG_Clean": ecg_cleaned,
"ECG_Rate": rate,
"ECG_Quality": quality})
signals = pd.concat([signals, instant_peaks], axis=1)
# Create info dict
info = rpeaks
info["sampling_rate"] = 1000
return signals, info
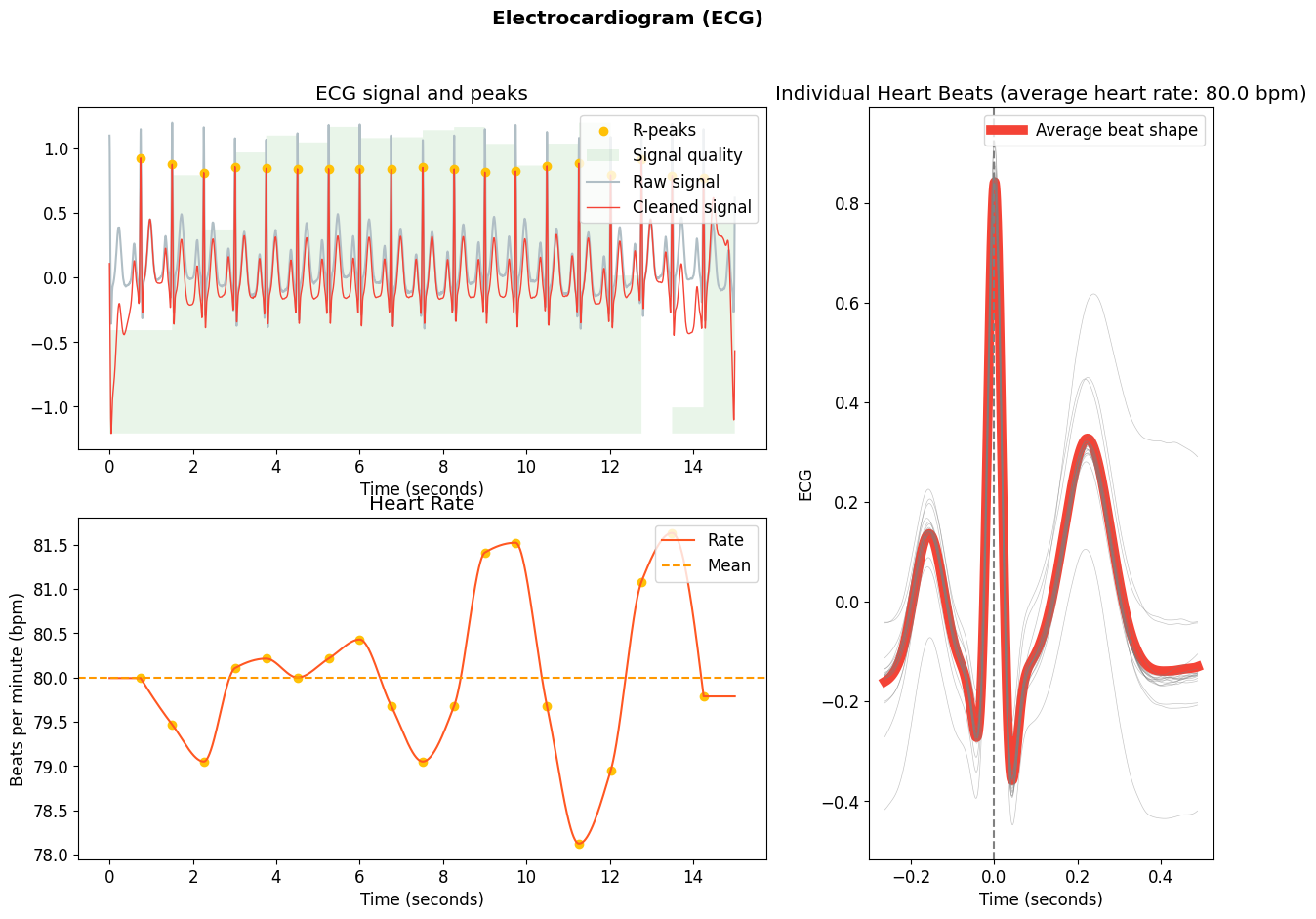
You can now use this function as you would do with ecg_process().
# Process the signal using previously defined function
signals, info = my_processing(ecg)
# Visualize
nk.ecg_plot(signals, info)
Changing the processing parameters#
Now, you might want to ask, why would you re-create the processing function? Well, it allows you to change the parameters of the inside as you please. Let’s say you want to use a specific cleaning method.
First, let’s look at the documentation for ecg_clean(), you can see that they are several different methods for cleaning which can be specified. The default is the NeuroKit method, however depending on the quality of your signal (and several other factors), other methods may be more appropriate. It is up to you to make this decision.
You can now change the methods as you please for each function in your custom processing function that you have written above:
# Define a new function
def my_processing(ecg_signal):
# Do processing
ecg_cleaned = nk.ecg_clean(ecg_signal, sampling_rate=1000, method="engzeemod2012")
instant_peaks, rpeaks, = nk.ecg_peaks(ecg_cleaned, sampling_rate=1000)
rate = nk.ecg_rate(rpeaks, sampling_rate=1000, desired_length=len(ecg_cleaned))
quality = nk.ecg_quality(ecg_cleaned, sampling_rate=1000)
# Prepare output
signals = pd.DataFrame({"ECG_Raw": ecg_signal,
"ECG_Clean": ecg_cleaned,
"ECG_Rate": rate,
"ECG_Quality": quality})
signals = pd.concat([signals, instant_peaks], axis=1)
# Create info dict
info = rpeaks
info["sampling_rate"] = 1000
return signals, info
Similarly, you can select a different method for the peak detection.
Customize even more!#
It is possible that none of these methods suit your needs, or that you want to test a new method. Rejoice yourself, as NeuroKit allows you to do that by providing what can be referred to as “low-level” functions.
For instance, you can rewrite the cleaning procedure by using the signal processing tools offered by NeuroKit:
def my_cleaning(ecg_signal, sampling_rate):
detrended = nk.signal_detrend(ecg_signal, order=1)
cleaned = nk.signal_filter(detrended,
sampling_rate=sampling_rate,
lowcut=2,
highcut=9,
method='butterworth')
return cleaned
You can use this function inside your custom processing written above:
# Define a new function
def my_processing(ecg_signal):
# Do processing
ecg_cleaned = my_cleaning(ecg_signal, sampling_rate=1000)
instant_peaks, rpeaks, = nk.ecg_peaks(ecg_cleaned, sampling_rate=1000)
rate = nk.ecg_rate(rpeaks, sampling_rate=1000, desired_length=len(ecg_cleaned))
quality = nk.ecg_quality(ecg_cleaned, sampling_rate=1000)
# Prepare output
signals = pd.DataFrame({"ECG_Raw": ecg_signal,
"ECG_Clean": ecg_cleaned,
"ECG_Rate": rate,
"ECG_Quality": quality})
signals = pd.concat([signals, instant_peaks], axis=1)
# Create info dict
info = rpeaks
info["sampling_rate"] = 1000
return signals, info
Congrats, you have created your own processing pipeline! Let’s see how it performs:
signals, info = my_processing(ecg)
nk.ecg_plot(signals, info)
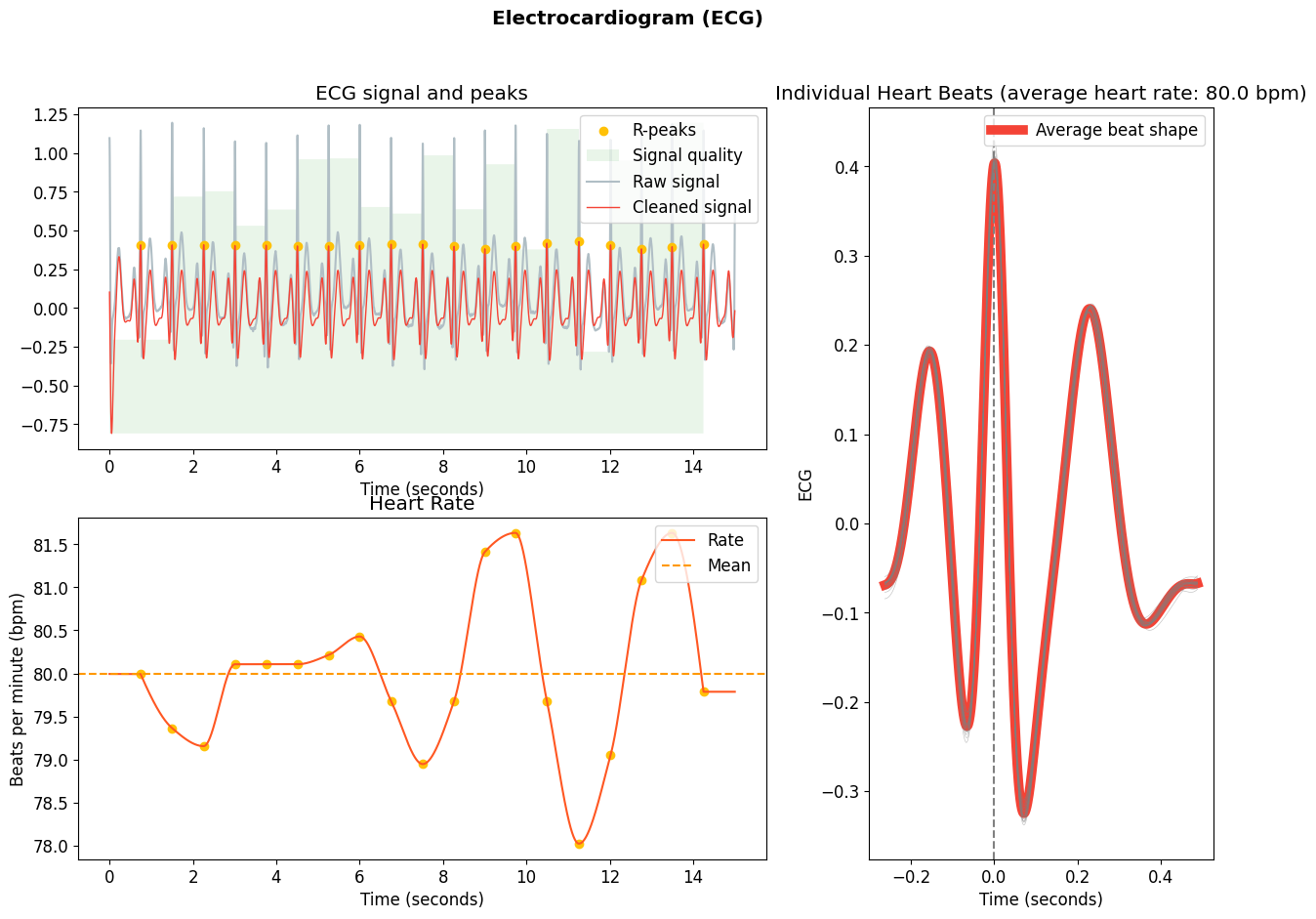
This doesn’t look bad :) Can you do better?
