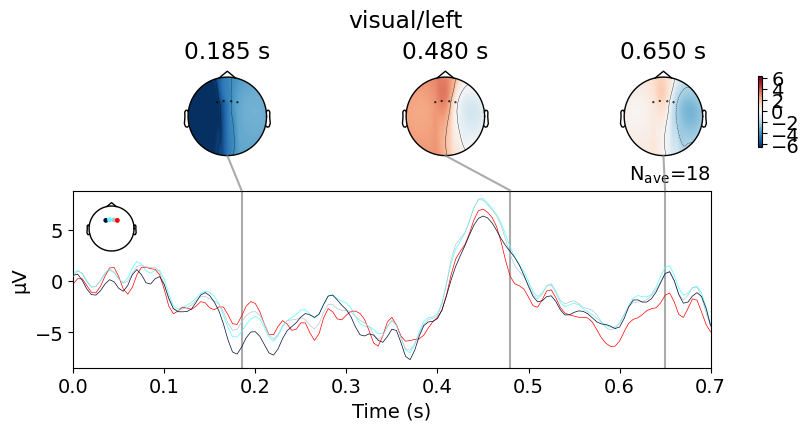
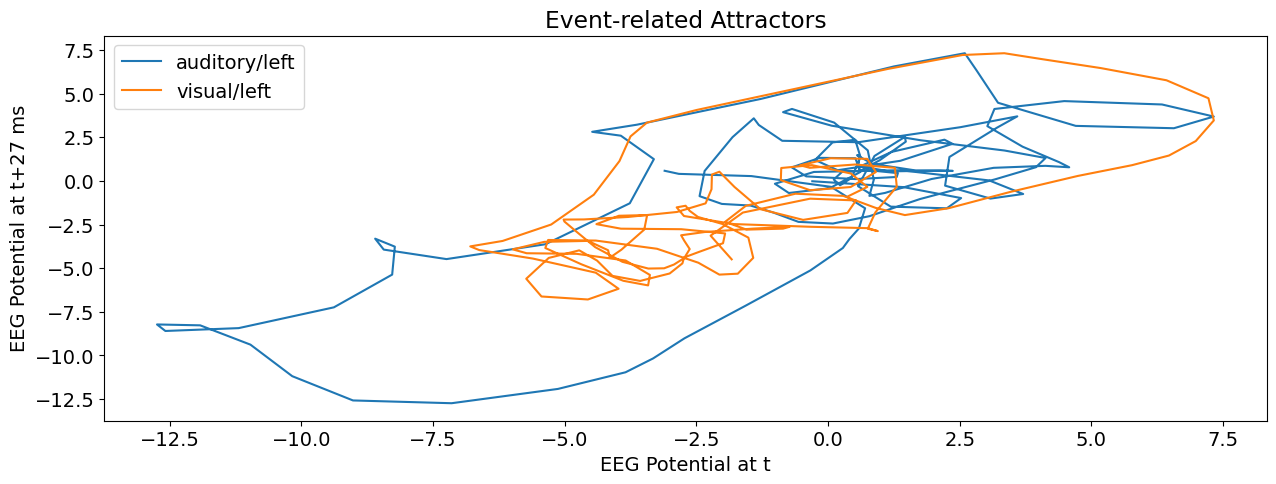
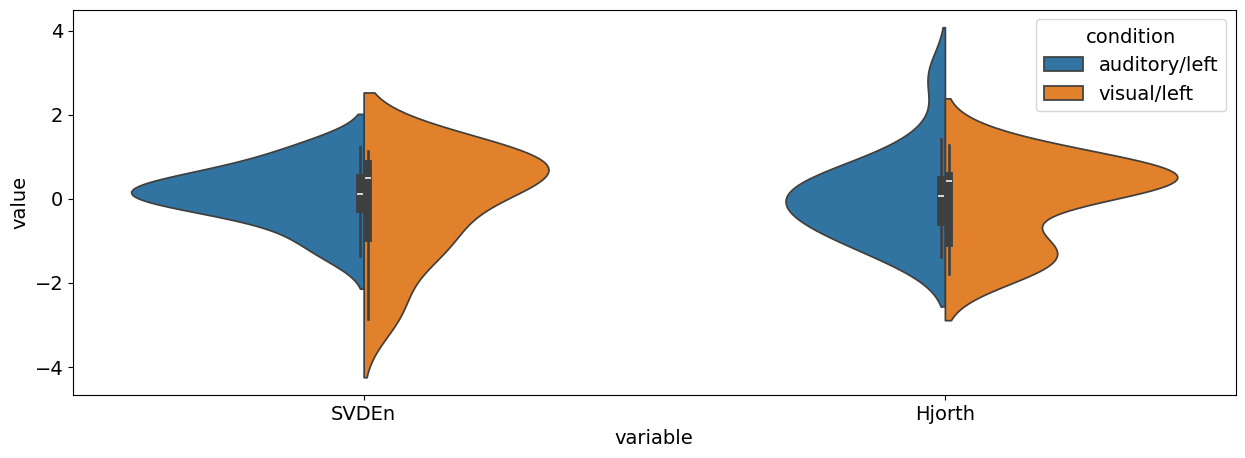
EEG Complexity Analysis#
In this example, we are going to apply complexity analysis to EEG data. Useful reads include:
This example below can be referenced by citing the package.
# Load packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats
import mne
import neurokit2 as nk
Data Preprocessing#
Load and format the EEG data of one participant following this MNE example.
# Load raw EEG data
raw = nk.data("eeg_1min_200hz")
# Find events and map them to a condition (for event-related analysis)
events = mne.find_events(raw, stim_channel='STI 014', verbose=False)
event_dict = {'auditory/left': 1, 'auditory/right': 2, 'visual/left': 3,
'visual/right': 4, 'face': 5, 'buttonpress': 32}
# Select only relevant channels
raw = raw.pick('eeg', verbose=False)
# Store sampling rate
sampling_rate = raw.info["sfreq"]
raw = raw.filter(l_freq=0.1, h_freq=40,verbose=False)
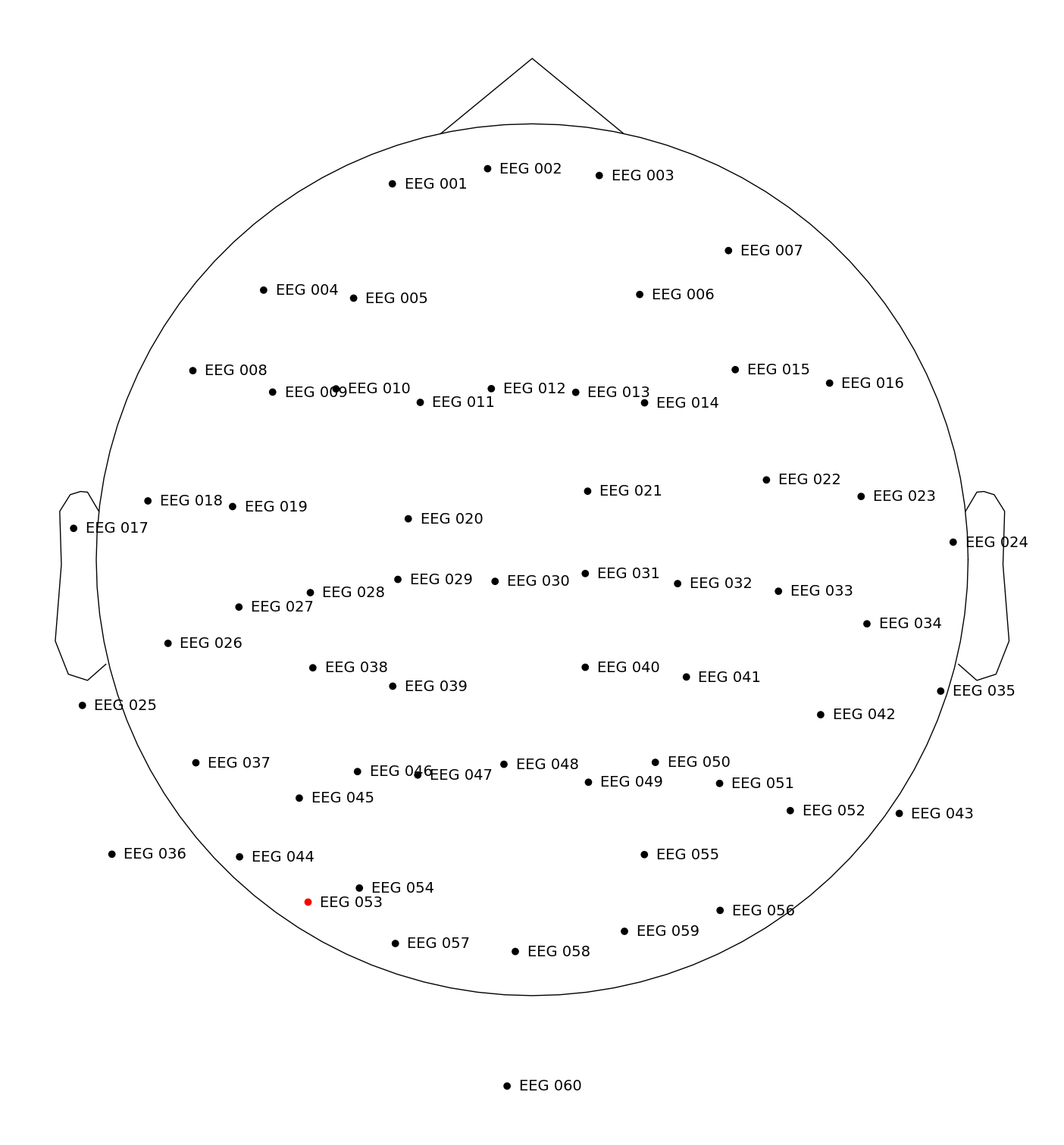
fig = raw.plot_sensors(show_names=True)